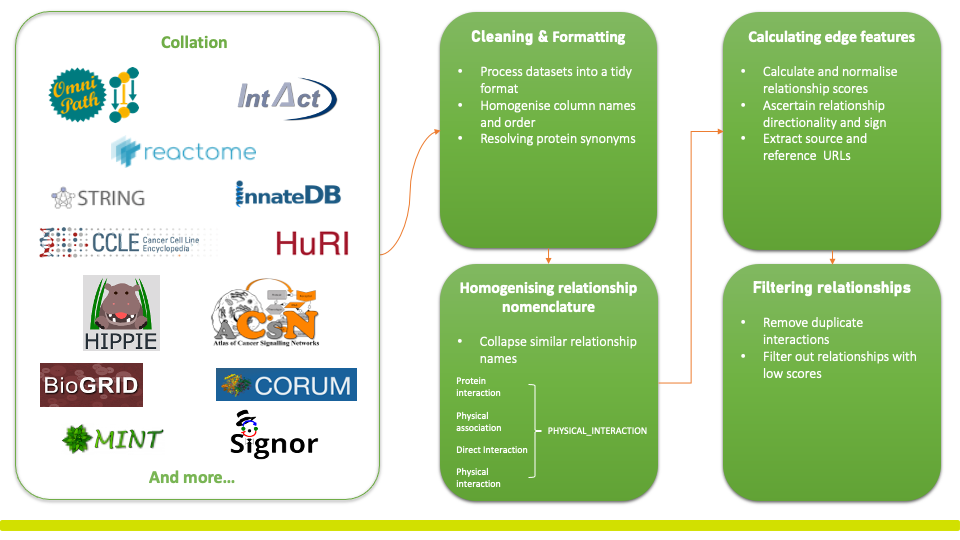
| Database | Short Description | Number of Entries | Scoring System |
|---|
| BioPlex | A protein-protein interaction database containing ~120000 interactions among ~15000 proteins. Profiling protein-protein interactions in human cells by affinity-purification mass spectrometry in human 293T and HCT116 cells. | 104,485 | Identifying high-confidence interreacting proteins (HCIP's) using the Normalized Weighted D-Score and Z-Score. Interactions scores of at least 0.75 are considered HCIP's. |
| Omnipath | A database of molecular biology prior knowledge combining data from >100 resources containing protein-protein and gene regulatory interactions, enzyme-PTM relationships, protein complexes, protein annotations and intercellular communication. | 202 | Developed based on the number of publications reporting an interaction. |
| STRING | A database of known and predicted protein-protein interactions. Interactions include direct (physical) and indirect (functional) associations. They stem from computational prediction, from knowledge transfer between organisms and from interactions aggregated from other (primary) databases. | 37081 | Indictor of confidence, i.e. how likely STRING judges an interaction to be true, given all available evidence. All scores rank from 0 to 1 with 1 being the highest possible confidence. |
| CORUM | A comprehensive dataset of protein complexes for discoveries in systems biology, analyses of protein networks and protein complex-associated diseases. | 1,636 | Developed based on the number of publications reporting an interaction. |
| HuRI | A protein-protein interaction database containing ~17500 interactions detected by high throughput yeast two-hybrid screens. This database also includes interactions of high quality extracted from literature. | 10,237 | Indicator of confidence to rank human binary protein interactions identified in systematic screens based on their biophysical quality. |
| Reactome | An open-source manually curated and peer-reviewed pathway database. Providing a bioinformatic tool for visualising, interpreting and analysing pathway knowledge to support genome analysis and systems biology. Entities including proteins, complexes, and small molecules form a network of biological interactions. | 20,630 | Developed based on the number of publications reporting an interaction. |
| XLinkDB | A protein-protein interaction database integrating tools for network analysis. Incorporating cross-linking data and model scoring scripts to identify interactions and analyse structural landscape predicted by crosslinking with mass spectrometry. | 3393 | Indictor of confidence, estimating the reliability of the interaction, given the available experimental evidence based on a heuristic integration. |
| BioGRID | A biomedical interaction repository with data compiled through comprehensive curation efforts with 2,604,369 protein and genetic interactions, 30,725 chemical interactions and 1,128,339 post translational modifications from major model organism species. | 91917 | Developed based on the number of publications reporting an interaction. |
| Signor | A resource annotating experimental evidence about casual interactions between proteins and other entities of biological relevance. The curated data is displayed as signed directed graphs by a graph drawing tool. | 12,500 | A score to support the functional relevance of the considered relationship. Based on the number of annotated articles reporting work that support the interaction and relationship occurrence in the Human Protein Interactions file from Reactome. |
| MINT | A protein-protein interaction database integrating experimentally verified interactions mined from scientific literature by expert curators. | 1,129 | A score estimating the reliability of the interaction, given the available experimental evidence based on a heuristic integration. The value of 0.50 is evidence supporting association, which may be indirect between the two partners. |
| InnateDB | A publicly available database of genes, proteins, experimentally verified interactions and signalling pathways involved in the innate immune response of organisms. Integrating known interactions and pathways from public databases together with manually curated data into a centralised resource. | 1,896 | Developed based on the number of publications reporting an interaction. |
| HIPPIE | A protein-protein interaction database, providing confidence scored and functionally annotated human interactions. | 180 | Indicator of confidence based on the amount and reliability of evidence supporting the interaction. Calculated as a weighted sum of the number of studies in which an interaction was detected and the number and quality of experimental techniques used to measure the interaction. |
| ACSN | A multi-scale resource of biological maps depicting molecular processes in cancer cells. Depicts molecular mechanisms at the level of biochemical interaction, forming a seamless network of >8,000 reactions covering close to 3,000 proteins and 800 genes. | 13,019 | Developed based on the number of publications reporting an interaction. |
| IntAct | An open-source database and analysis tool for molecular interaction data derived from literature curation or direct user submission currently comprising of 118924 interactors and 750480 interactions. | 93 | A heuristic scoring system considering how the interaction was inferred, the interaction type and the number of publications reporting a scientific interaction. |
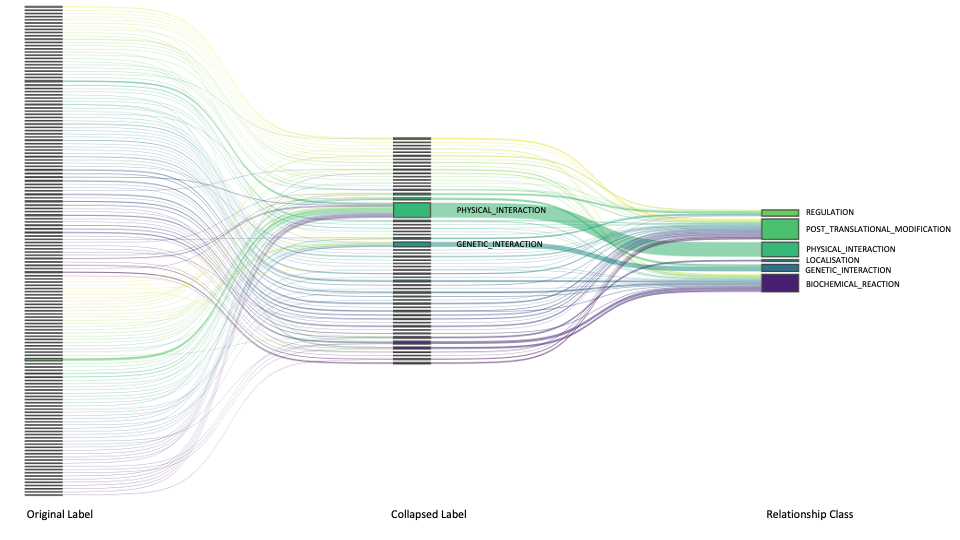
| Original Label | Collapsed Label | Relationship Class | Database |
|---|
| CATALYSIS | CATALYSIS | BIOCHEMICAL_REACTION | ACSN |
| UNKNOWN_CATALYSIS | CATALYSIS | BIOCHEMICAL_REACTION | ACSN |
| oxidoreductase activity electron transfer reaction | CATALYSIS | BIOCHEMICAL_REACTION | Reactome |
| nucleoside triphosphatase reaction | CATALYSIS | BIOCHEMICAL_REACTION | Reactome |
| phospholipase reaction | CATALYSIS | BIOCHEMICAL_REACTION | Reactome |
| catalytic activity | CATALYSIS | BIOCHEMICAL_REACTION | SIGNOR |
| cleavage reaction | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | Reactome |
| cleavage | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | SIGNOR |
| cleavage reaction | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | MINT |
| cleavage reaction | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | PINA |
| protein cleavage | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | PINA |
| CLEAVAGE_REACTION | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | Reactome |
| DNA_CLEAVAGE | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | Intact |
| rna cleavage | CLEAVAGE_REACTION | BIOCHEMICAL_REACTION | PINA |
| covalent binding | COVALENT_BINDING | BIOCHEMICAL_REACTION | PINA |
| covalent binding | COVALENT_BINDING | BIOCHEMICAL_REACTION | MINT |
| dephosphorylation reaction | DEPHOSPHORYLATION | BIOCHEMICAL_REACTION | PINA |
| dephosphorylation reaction | DEPHOSPHORYLATION | BIOCHEMICAL_REACTION | MINT |
| disulfide bond | DISULFIDE_BOND | BIOCHEMICAL_REACTION | MINT |
| disulfide bond | DISULFIDE_BOND | BIOCHEMICAL_REACTION | PINA |
| enzymatic reaction | ENZYMATIC_REACTION | BIOCHEMICAL_REACTION | Reactome |
| ENZYMATIC_REACTION | ENZYMATIC_REACTION | BIOCHEMICAL_REACTION | Reactome |
| enzymatic reaction | ENZYMATIC_REACTION | BIOCHEMICAL_REACTION | PINA |
| enzymatic reaction | ENZYMATIC_REACTION | BIOCHEMICAL_REACTION | MINT |
| oxidoreductase activity electron transfer reaction | ENZYMATIC_REACTION | BIOCHEMICAL_REACTION | MINT |
| KNOWN_TRANSITION_OMITTED | FOLDING_REACTION | BIOCHEMICAL_REACTION | ACSN |
| guanine nucleotide exchange factor | GTPASE_REACTION | BIOCHEMICAL_REACTION | SIGNOR |
| gtpase-activating protein | GTPASE_REACTION | BIOCHEMICAL_REACTION | SIGNOR |
| gtpase reaction | GTPASE_REACTION | BIOCHEMICAL_REACTION | Reactome |
| guanine nucleotide exchange factor reaction | GTPASE_REACTION | BIOCHEMICAL_REACTION | MINT |
| gtpase reaction | GTPASE_REACTION | BIOCHEMICAL_REACTION | PINA |
| HETERODIMER_ASSOCIATION | HETERODIMER_ASSOCIATION | BIOCHEMICAL_REACTION | ACSN |
| INHIBITION | INHIBITION | BIOCHEMICAL_REACTION | ACSN |
| isomerase reaction | ISOMERASE REACTION | BIOCHEMICAL_REACTION | Reactome |
| oxidation | OXIDATION | BIOCHEMICAL_REACTION | SIGNOR |
| palmitoylation | PALMITOYLATION | BIOCHEMICAL_REACTION | SIGNOR |
| phosphorylation reaction | PHOSPHORYLATION | BIOCHEMICAL_REACTION | MINT |
| PHOSPHORYLATION_REACTION | PHOSPHORYLATION | BIOCHEMICAL_REACTION | Intact |
| proline isomerization reaction | PROLINE_ISOMERIZATION | BIOCHEMICAL_REACTION | MINT |
| relocalization | RELOCALISATION | BIOCHEMICAL_REACTION | SIGNOR |
| COMPLEX_EXPANSION | SHARES_COMPLEX | BIOCHEMICAL_REACTION | ACSN |
| SHARE_COMPLEX | SHARES_COMPLEX | BIOCHEMICAL_REACTION | Corum |
| UNKNOWN_TRANSITION | STRUCTURAL_CHANGE | BIOCHEMICAL_REACTION | ACSN |
| STATE_TRANSITION | STRUCTURAL_CHANGE | BIOCHEMICAL_REACTION | ACSN |
| destabilization | STRUCTURAL_CHANGE | BIOCHEMICAL_REACTION | SIGNOR |
| sulfurtransfer reaction | SULFUR_TRANSFER | BIOCHEMICAL_REACTION | Reactome |
| transglutamination reaction | TRANSGLUTAMINATION | BIOCHEMICAL_REACTION | PINA |
| translation regulation | TRANSLATIONAL_REGULATION | BIOCHEMICAL_REACTION | SIGNOR |
| TRANSLATION | TRANSLATIONAL_REGULATION | BIOCHEMICAL_REACTION | ACSN |
| genetic interaction | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | PINA |
| Negative Genetic | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Dosage Lethality | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| NEGATIVE_GENETIC | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| SYNTHETIC_LETHALITY | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| DOSAGE_RESCUE | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| suppressive genetic interaction defined by inequality | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | PINA |
| Phenotypic Suppression | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Synthetic Growth Defect | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Dosage Growth Defect | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| synthetic genetic interaction defined by inequality | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | PINA |
| SYNTHETIC_GROWTH_DEFECT | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| PHENOTYPIC_SUPPRESSION | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| Synthetic Lethality | NEGATIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| additive genetic interaction defined by inequality | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | PINA |
| Positive Genetic | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Phenotypic Enhancement | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Synthetic Rescue | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| Dosage Rescue | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGRID |
| PHENOTYPIC_ENHANCEMENT | POSITIVE_GENETIC_INTERACTION | GENETIC_INTERACTION | BioGrid |
| colocalization | COLOCALIZATION | LOCALIZATION | PINA |
| proximity | COLOCALIZATION | LOCALIZATION | MINT |
| colocalization | COLOCALIZATION | LOCALIZATION | MINT |
| PROXIMITY | COLOCALIZATION | LOCALIZATION | Intact |
| COLOCALIZATION | COLOCALIZATION | LOCALIZATION | Intact |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | IID |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | STRING |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioPlex |
| physical association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | HuRI |
| physical association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Reactome |
| physical association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PINA |
| POST_TRANSLATIONAL_REGULATION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Omnipath |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PIPs |
| Affinity Capture-Western | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Affinity Capture-MS | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Reconstituted Complex | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| direct interaction | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PINA |
| association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | MINT |
| physical association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | MINT |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | XLinkDB |
| Two-hybrid | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Biochemical Activity | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| association | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PINA |
| direct interaction | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | MINT |
| Proximity Label-MS | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| binding | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | SIGNOR |
| Co-localization | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | InnateDB |
| Co-crystal Structure | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Protein-peptide | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Co-purification | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Co-fractionation | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Affinity Capture-Luminescence | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| FRET | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| PHYSICAL_ASSOCIATION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Reactome |
| Far Western | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| Affinity Capture-RNA | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| PCA | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| ASSOCIATION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Intact |
| Protein-RNA | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGRID |
| PHYSICAL_ASSOCIATION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Intact |
| PROTEIN_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | HIPPIE |
| PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | BioGrid |
| PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | STRING |
| DIRECT_INTERACTION | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | Intact |
| physical interaction | PHYSICAL_INTERACTION | PHYSICAL_INTERACTION | PINA |
| acetylation reaction | ACETYLATION_REACTION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| acetylation | ACETYLATION_REACTION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| acetylation reaction | ACETYLATION_REACTION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| acetylation reaction | ACETYLATION_REACTION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| adp ribosylation reaction | ADP_RIBOSYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| adp ribosylation reaction | ADP_RIBOSYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| ADP-ribosylation | ADP_RIBOSYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| amidation reaction | AMIDATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| demethylation | CARBOXYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| carboxylation reaction | CARBOXYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| de-ADP-ribosylation reaction | DE-ADP-RIBOSYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| deacetylation reaction | DEACETYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| deacetylation reaction | DEACETYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| deacetylation reaction | DEACETYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| deacetylation | DEACETYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| deglycosylation | DEGLYCOSYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| demethylation reaction | DEMETHYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| carboxylation | DEMETHYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| demethylation reaction | DEMETHYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| deneddylation reaction | DENEDDYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| deneddylation reaction | DENEDDYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| deneddylation reaction | DENEDDYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| dephosphorylation | DEPHOSPHORYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| dephosphorylation reaction | DEPHOSPHORYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| desumoylation | DESUMOYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| deubiquitination | DEUBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| deubiquitination reaction | DEUBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| deubiquitination reaction | DEUBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| deubiquitination reaction | DEUBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| glycosylation reaction | GLYCOSLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| glycosylation | GLYCOSLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| hydroxylation reaction | HYDROXYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| hydroxylation | HYDROXYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| hydroxylation reaction | HYDROXYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| isomerization | ISOMERIZATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| lipidation | LIPIDATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| methylation | METHYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| methylation reaction | METHYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| methylation reaction | METHYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| monoubiquitination | MONOUBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| neddylation | NEDDYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| palmitoylation reaction | PALMITOYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| phosphopantetheinylation | PHOSPHOPANTETHEINYLATION | POST_TRANSLATIONAL_MODIFICATION | Reactome |
| phosphorylation | PHOSPHORYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| phosphorylation reaction | PHOSPHORYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| post transcriptional regulation | POST_TRANSLATIONAL_REGULATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| post translational modification | POST_TRANSLATIONAL_REGULATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| s-nitrosylation | S-NITROSYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| sumoylation | SUMOYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| sumoylation reaction | SUMOYLATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| sumoylation reaction | SUMOYLATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| trimethylation | TRIMETHYLATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| tyrosination | TYROSINATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| ubiquitination | UBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| polyubiquitination | UBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | SIGNOR |
| ubiquitination reaction | UBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | PINA |
| ubiquitination reaction | UBIQUITINATION | POST_TRANSLATIONAL_MODIFICATION | MINT |
| DISSOCIATION | DISSOCIATION | REGULATION | ACSN |
| UNKNOWN_INHIBITION | INHIBITION | REGULATION | ACSN |
| NEGATIVE_INFLUENCE | NEGATIVE_REGULATION | REGULATION | ACSN |
| NUCLEOSIDE_TRIPHOSPHATASE_REACTION | NEGATIVE_REGULATION | REGULATION | Reactome |
| chemical inhibition | NEGATIVE_REGULATION | REGULATION | SIGNOR |
| UNKNOWN_NEGATIVE_INFLUENCE | NEGATIVE_REGULATION | REGULATION | ACSN |
| transcriptional repression | NEGATIVE_TRANSCRIPTIONAL_REGULATION | REGULATION | SIGNOR |
| PHYSICAL_STIMULATION | POSITIVE_REGULATION | REGULATION | ACSN |
| TRIGGER | POSITIVE_REGULATION | REGULATION | ACSN |
| POSITIVE_INFLUENCE | POSITIVE_REGULATION | REGULATION | ACSN |
| chemical activation | POSITIVE_REGULATION | REGULATION | SIGNOR |
| UNKNOWN_POSITIVE_INFLUENCE | POSITIVE_REGULATION | REGULATION | ACSN |
| neddylation reaction | POST_TRANSLATIONAL_REGULATION | REGULATION | MINT |
| MODULATION | REGULATION | REGULATION | ACSN |
| TRANSCRIPTIONAL_REGULATION | TRANSCRIPTIONAL_REGULATION | REGULATION | Omnipath |
| transcriptional regulation | TRANSCRIPTIONAL_REGULATION | REGULATION | SIGNOR |
| TRANSCRIPTION | TRANSCRIPTIONAL_REGULATION | REGULATION | ACSN |